The Stelling Lab uses solution state bioanalytical methods, with a focus on vibrational spectroscopy, to probe molecular interactions in proteins and nucleic acids.
Unraveling DNA structure
(A) Measuring hydrogen bond strength in DNA duplexes with IR spectroscopy
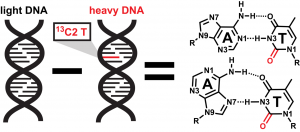
Site-specific incorporation of heavy atoms into base pairs in DNA duplexes will be used to measure the strength of their hydrogen bonds and to determine how this is impacted by sequence context.
(B) Detection of A-T Hoogsteen base pairs in DNA:protein complexes with IR and Raman spectroscopy
Recent studies have exposed a new layer of dynamic complexity in DNA that can potentially redefine our basic view of the structure of DNA in vivo. NMR studies have shown that in canonical duplex DNA, G-C and A-T Watson-Crick base pairs exist in dynamic equilibrium with alternative Hoogsteen base pairs in which the purine flips 180º to form a new set of hydrogen bonds. There are now several documented examples in which Hoogsteen base pairs play crucial roles in DNA based transactions including in damage induction, accommodation, and repair, replication, and sequence-specific DNA-protein recognition. Despite their growing importance, visualizing Hoogsteen base pairs in protein-DNA complexes remains an outstanding challenge in structural biology principally as they are very challenging to distinguish from Watson-Crick base pairs using X-ray crystallography. Indeed, a preliminary analysis of local DNA B-factors in high resolution nucleosome crystal structure reveals that most of the DNA bases are not well defined. The presence of Hoogsteen base pairs can account for these higher B-factors, and indicates a need for solution-based methods sensitive to fine molecular details for their detection. This project builds on our previous work which developed robust infrared (IR) methods for detecting the unambiguous signals of G-C+ Hoogsteen base pairs in a DNA:protein complex in solution. However, it is A-T Hoogsteen base pairs that are thought to occur more frequently in cells, as their populations can be dramatically elevated in DNA when the duplex structure is perturbed by drug or protein binding due to their low energy barrier for formation. In this project, we will develop new methods for validating the complex IR signals that arise from A-T Hoogsteen base pairs. We will then apply these methods to test the hypothesis that the high B-factor regions of DNA bound to the nucleosome core particle is enriched in Hoogsteen base pairs.
Analysis of S-adenosyl-l-methionine conformation
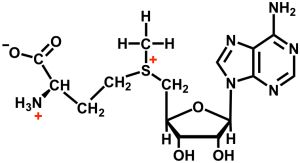
SAM is an essential cofactor for cellular methyl transfer reactions that control the placement of methyl groups on proteins and nucleic acids. These methylation events are critical for cellular function and epigenetic regulation, and their disregulation is a major driver in human cancers. Due to their importance in disease, SAM dependent methyltranserases are important drug targets. Yet, specificity remains a challenge and a deeper understanding of SAM’s conformation when bound to enzymes is needed to help guide the discovery of high affinity ligands.
This project will use solution state structural methods sensitive to molecular structure (IR and Raman spectroscopy) to detect the conformation of SAM when bound to active sites, and determine enzyme-SAM interactions important for SAM binding. This information on the SAM:protein interface will be used to design small molecule probes that exploit the interface to increase ligand specificity and affinity.
